Published:2021-01-04 01:00:00.0
The Virtual Brain is an open-source, multi-scale, multi-modal brain simulation platform. In this lesson, you get introduced to brain simulation in general and to The Virtual brain in particular. Prof. Ritter will present the newest approaches for clinical applications of The Virtual brain - that is, for stroke, epilepsy, brain tumors and Alzheimer’s disease - and show how brain simulation can improve diagnostics, therapy and understanding of neurological disease.
Topics covered in this lesson by Petra Ritter
- Introduction to non-linear systems theory
- Neural masses and mean field theory
- Multi-scale brain modeling
- Excitation and inhibition as crucial factors for brain dynamics
- Simulation of stroke and prediction of clinical outcome
- The virtual epileptic patient - towards improved neurosurgery in epilepsy
- Simulation of brain tumors
- Prediction of cognitive scores in Alzheimer’s disease by brain simulation
- Linking molecular features of neurodegeneration with multi-scale modeling in Alzheimer’s disease
- Improving the functionality of brain computer interfaces
byMichael Burgstahler
Published:2021-01-03 01:00:00.0
This lecture briefly introduces The Virtual Brain (TVB), a multi-scale, multi-modal neuroinformatics platform for full brain network simulations using biologically realistic connectivity, as well as its potential neuroscience applications: for example with epilepsy.
Topics covered in this lesson by Petra Ritter
- Introduction to TVB
- Motivations and achievements of TVB
- Presentation of BrainModes App
- Outlook of multi-scale brain network
byMichael Burgstahler
Published:2021-01-02 01:00:00.0
The practical usage of The Virtual brain in its graphical user interface and via python scripts is introduced. In the graphical user interface, you are guided through its data repository, simulator, phase plane exploration tool, connectivity editor, stimulus generator and the provided analyses.
The implemented iPython notebooks of TVB are presented, and since they are public, can be used for further exploration of The Virtual brain.
Topics covered in this lesson by Paul Triebkorn
- Introduction to the GUI
- Data repository
- Connectivity editor
- Simulator
- Phase plane exploration tool
- Exploring simulation results
- Generating a brain stimulus
- Parameter space exploration
- Brain function visualizers
- iPython notebooks and python scripting with TVB
byMichael Burgstahler
Published:2021-01-01 01:00:00.0
This lecture presents the Graphical (GUI) and Command Line (CLI) User Interface of TVB. Alongside with the speakers, explore and interact with all means necessary to generate, manipulate and visualize connectivity and network dynamics.
Topics covered in this lesson by Paula Popa & Mihai Andrei
- Setting up the Graphical User Interface (GUI)
- Workflow of TVB and its major activities:
- Project: data management
- Connectivity: edit a connectivity matrix
- Simulator
- Exploring a model behavior
- Set up a region model
- Launch a simulator
- Perform a parameter sweep
- Stimulus
- Visualizers
- Access Jupyter Notebooks of TVB
byMichael Burgstahler
Published:2020-12-31 01:00:00.0
This hands-on focuses on a brief introduction to the GUI of TVB. You will visualize a structural connectome and use it for simulation. The local neural mass model will be explored through the phase plane viewer and a parameter space exploration will be performed to observe different dynamics of the large-scale brain model.
Topics covered in this lesson by Paul Triebkorn
- TVB graphical user interface
- Connectome viewer
- Phase plane viewer
- Parameter space exploration
- Simulation visualization
byMichael Burgstahler
Published:2020-12-30 01:00:00.0
This tutorial on how to simulate using TVB is part of the TVB Node 10 series, a 4 day workshop dedicated to learning about The Virtual Brain, brain imaging, brain simulation, personalised brain models, TVB use cases, etc. TVB is a full brain simulation platform.
Topics covered in this lesson by Paul Triebkorn
- Step by step guide to performing a simulation with TVB
byMichael Burgstahler
Published:2020-12-29 01:00:00.0
The concept of neural masses, an application of mean field theory, is introduced as a possible surrogate for electrophysiological signals in brain simulation. The mathematics of neural mass models and their integration to a coupled network are explained.
Bifurcation analysis is presented as an important technique in the understanding of non-linear systems and as a fundamental method in the design of brain simulations.
Finally, the application of the described mathematics is demonstrated in the exploration of brain stimulation regimes.
Topics covered in this lesson by Andreas Spiegler
- Mathematical description of neural masses
- Transfer functions in local brain dynamics
- Mathematical implementation of structural connectivity
- The Jansen-Rit model
- Bifurcation analysis
- Equilibrium manifolds
- Dynamics of brain stimulation
byMichael Burgstahler
Published:2020-12-28 01:00:00.0
This lecture introduces the theoretical background and foundations that led to the development of TVB, the architecture and features of its major software components.
Topics covered in this lesson by Randy McIntosh
- Motivation for The Virtual Brain
- The Virtual Brain Platform
- Architecture and access
- Dynamics
- Geometry & Connectivity
- Simulations & Data Generation
- The Virtual Brain applied
byMichael Burgstahler
Published:2020-12-27 01:00:00.0
-
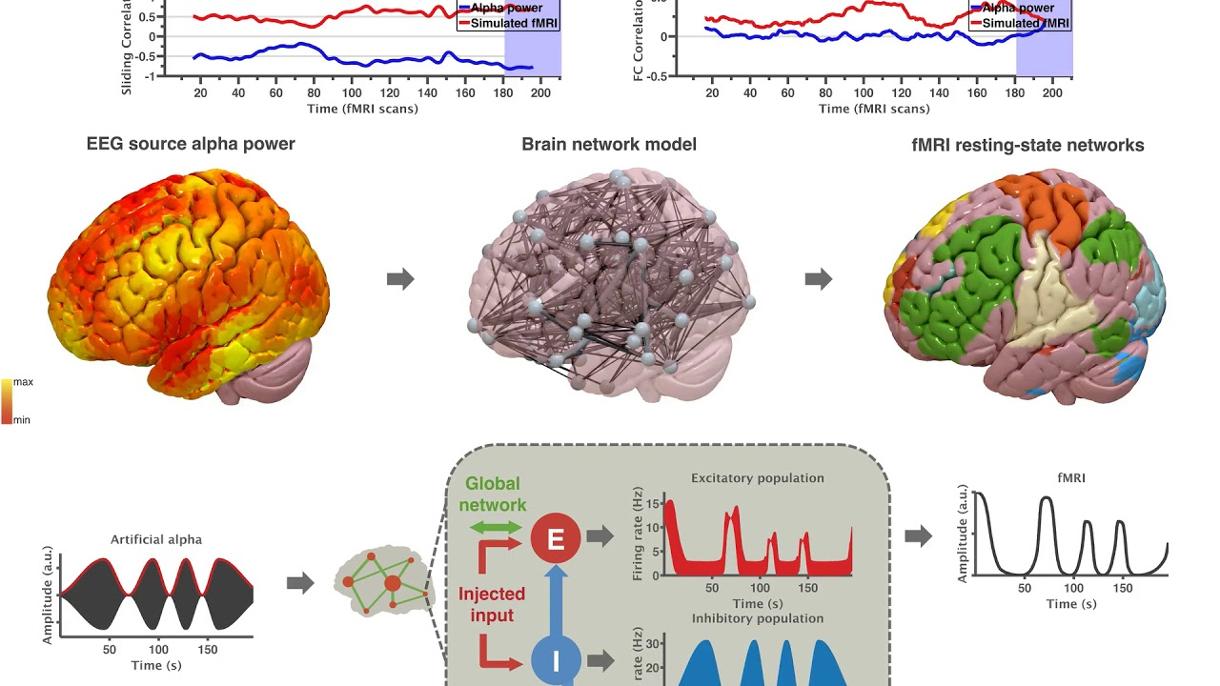
Related publication
Inferring multi-scale neural mechanisms with brain network modelling, published in eLife, January 2018, by Michael Schirner, Anthony Randal McIntosh, Viktor Jirsa, Gustavo Deco, Petra Ritter
doi: 10.7554/eLife.28927
Abstract
The neurophysiological processes underlying non-invasive brain activity measurements are incompletely understood. Here, we developed a connectome-based brain network model that integrates individual structural and functional data with neural population dynamics to support multi-scale neurophysiological inference.
Simulated populations were linked by structural connectivity and, as a novelty, driven by electroencephalography (EEG) source activity. Simulations not only predicted subjects' individual resting-state functional magnetic resonance imaging (fMRI) time series and spatial network topologies over 20 minutes of activity, but more importantly, they also revealed precise neurophysiological mechanisms that underlie and link six empirical observations from different scales and modalities:
- (1) resting-state fMRI oscillations,
- (2) functional connectivity networks,
- (3) excitation-inhibition balance,
- (4, 5) inverse relationships between α-rhythms, spike-firing and fMRI on short and long time scales,
- and (6) fMRI power-law scaling.
These findings underscore the potential of this new modelling framework for general inference and integration of neurophysiological knowledge to complement empirical studies.
More resources for this lesson
GitHub repository with code
byMichael Burgstahler
Published:2020-12-26 01:00:00.0
This presentation on population models and phase plane is part of the TVB Node 10 series, a 4 day workshop dedicated to learning about The Virtual Brain, brain imaging, brain simulation, personalised brain models, TVB use cases, etc... TVB is a full brain simulation platform.
Topics covered in this lesson by Dr. Michael Schirner
- What is a population model
- What is phase plane
- Constructing a StefanescuJirsa population model
byMichael Burgstahler
