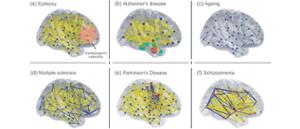
Simulating the human brain is the holy grail of neuroscience – offering a pioneering tool for understanding how our brain works and how to deal with its disorders like stroke, epilepsy or neurodegenerative diseases like Alzheimer’s or Parkinson’s.
While large-scale research initiatives simulate neurons and small brain regions at the cellular level on massively parallel hardware, they are still years away from clinical applications.
The Virtual Brain (TVB) takes a different approach and reduces complexity on the micro level to attain the macro organization: a TVB model of a patient's brain generates sufficiently accurate EEG, MEG, BOLD and SEEG signals by reducing the complexity millionfold through methods from statistical physics. The key is TVB’s hybrid approach of merging individual anatomy from brain imaging data with state-of-the-art mathematical modeling.
In a nutshell:
you can simulate a human brain with TVB right now, for free and with access to the source code – on average MacBooks, faculty Linux servers or supercomputers like Piz Daint in CSCS Lugano.
Since its early beginnings as network equations scribbled on a bar coaster in 2008, TVB has gained wide recognition – demonstrated by regular invitations to participate at neuroinformatics events such as INCF conferences, and workshops dedicated to High Performance Computing (HPC) (such as organized by NSG).
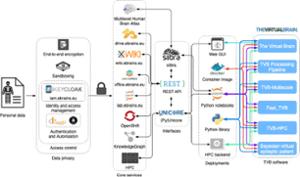
Today, TVB is a reference tool for full-brain simulation. TVB serves as the simulation engine in the world’s first clinical trial on predictive brain modeling in epilepsy surgery (EPINOV). TVB is also the choice of full-brain simulator in Europe’s digital research infrastructure EBRAINS and directly links to other large-scale neuroinformatics efforts such as the Allen Institute’s Mouse atlas or the Human Brain Project (HBP).
A large, growing user base of clinicians and scientists are working every day with TVB, citing its scientific groundwork in well over 100 peer-reviewed publications. Dedicated research facilities at Charité in Berlin, AMU in Marseille and Baycrest in Toronto have constructed and simulated almost 1000 individual, Connectome-based brain network models and published their findings, clocking in upwards of 10 million CPU core hours.